TransUNet:用Transformer为医学图像分割构造强力编码器.
在医学图像分割领域,Unet已经取得了很优秀的效果。但是,CNN结构并不擅长建立远程信息连接,也就是CNN结构的感受野有限。尽管可以通过堆叠CNN结构、使用空洞卷积等方式增加感受野,但也会引入包括但不限于卷积核退化、空洞卷积造成的栅格化等问题,导致最终效果受限。
Self-Attention机制可以不限距离的建立远程连接,突破了CNN模型感受野不足的问题。本文作者设计了TransUnet结构,结合了UNet模型和Transformer,具体地,前三层是CNN-based,但是最后一层是Transformer-based。也就是把Unet的encoder最后一层换成了Transformer模型。
TransUnet只将编码器其中一部分换成Transformer也是有它自己的考虑。虽然Transformer能够获得到全局的感受野,但是在细节特征的处理上存在缺陷。实验表明Transformer对于局部的细节分割是有缺陷的,而CNN反而是得益于其局部的感受野,能够较为精确恢复细节特征。因此TransUnet模型只替换了最后一层,而这一层则更多关注全局信息,这是Transformer擅长的,至于浅层的细节识别任务则由CNN来完成。
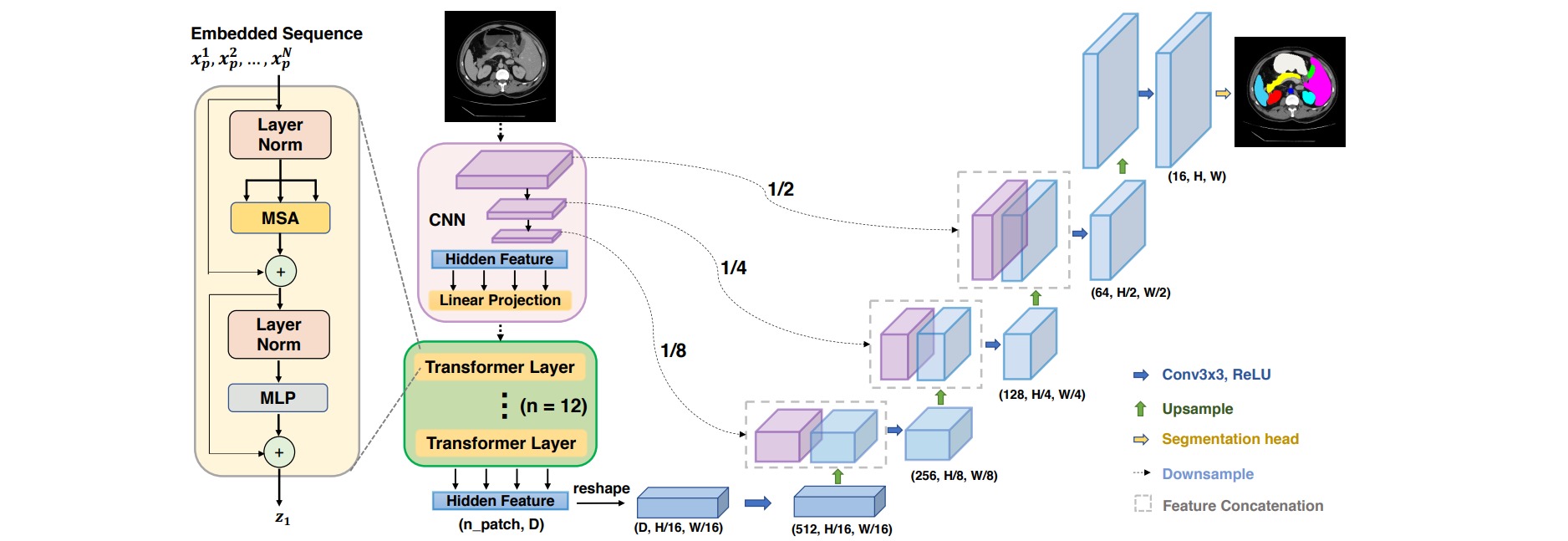
Encoder部分主要由ResNet50和Vit组成,在ResNet50部分,取消掉stem_block结构中的4倍下采样,保留前三层模型结构,这三层都选择两倍下采样,其中最后一层的输出作为Vit的输入。最后一层则是Vit结构,也就是12层Transformer Layer;作者把该encoder叫做R50-ViT。
class TransUnetEncoder(nn.Module):
def __init__(self, **kwargs):
super(TransUnetEncoder, self).__init__()
self.R50 = ResNet.resnet50()
self.Vit = ViT(image_size = (64, 64), patch_size = 16, channels = 256, dim = 512, depth = 12, heads = 16, mlp_dim = 1024, dropout = 0.1, emb_dropout = 0.1)
def forward(self, x):
x1, x2, x3 = self.R50(x)
x4 = self.Vit(x3)
return [x1, x2, x3, x4]
decoder结构很简单,还是典型的skip-connection和upsample结合。
class TransUnetDecoder(nn.Module):
def __init__(self, out_channels=64, **kwargs):
super(TransUnetDecoder, self).__init__()
self.decoder1 = nn.Sequential(
nn.Conv2d(out_channels//4, out_channels//4, 3, padding=1),
nn.BatchNorm2d(out_channels//4),
nn.ReLU()
)
self.upsample1 = nn.Sequential(
nn.UpsamplingBilinear2d(scale_factor=2),
nn.Conv2d(out_channels, out_channels//4, 3, padding=1),
nn.BatchNorm2d(out_channels//4),
nn.ReLU()
)
self.decoder2 = nn.Sequential(
nn.Conv2d(out_channels*2, out_channels, 3, padding=1),
nn.BatchNorm2d(out_channels),
nn.ReLU()
)
self.upsample2 = nn.Sequential(
nn.UpsamplingBilinear2d(scale_factor=2),
nn.Conv2d(out_channels*2, out_channels, 3, padding=1),
nn.BatchNorm2d(out_channels),
nn.ReLU()
)
self.decoder3 = nn.Sequential(
nn.Conv2d(out_channels*4, out_channels*2, 3, padding=1),
nn.BatchNorm2d(out_channels*2),
nn.ReLU()
)
self.upsample3 = nn.Sequential(
nn.UpsamplingBilinear2d(scale_factor=2),
nn.Conv2d(out_channels*4, out_channels*2, 3, padding=1),
nn.BatchNorm2d(out_channels*2),
nn.ReLU()
)
self.decoder4 = nn.Sequential(
nn.Conv2d(out_channels*8, out_channels*4, 3, padding=1),
nn.BatchNorm2d(out_channels*4),
nn.ReLU()
)
self.upsample4 = nn.Sequential(
nn.UpsamplingBilinear2d(scale_factor=2),
nn.Conv2d(out_channels*8, out_channels*4, 3, padding=1),
nn.BatchNorm2d(out_channels*4),
nn.ReLU()
)
def forward(self, inputs):
x1, x2, x3, x4 = inputs
# b 512 H/8 W/8
x4 = self.upsample4(x4)
x = self.decoder4(torch.cat([x4, x3], dim=1))
x = self.upsample3(x)
x = self.decoder3(torch.cat([x, x2], dim=1))
x = self.upsample2(x)
x = self.decoder2(torch.cat([x, x1], dim=1))
x = self.upsample1(x)
x = self.decoder1(x)
return x
最后将Encoder和Decoder包装成TransUnet。
class TransUnet(nn.Module):
# 主要是修改num_classes
def __init__(self, num_classes=4, **kwargs):
super(TransUnet, self).__init__()
self.TransUnetEncoder = TransUnetEncoder()
self.TransUnetDecoder = TransUnetDecoder()
self.cls_head = nn.Conv2d(16, num_classes, 1)
def forward(self, x):
x = self.TransUnetEncoder(x)
x = self.TransUnetDecoder(x)
x = self.cls_head(x)
return x


